Abstract
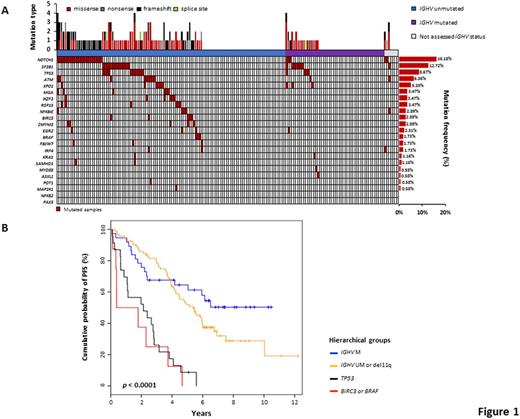
Background. The current shift of therapy of chronic lymphocytic leukemia (CLL) towards novel targeted agents mandates the recognition of molecular predictors to identify patients who can still benefit from chemoimmunotherapy and those who should instead be considered for novel targeted agents upfront. In the case of FCR (fludarabine, cyclophosphamide, rituximab), the IGHV mutation status and FISH karyotype stratify low-risk patients carrying mutated IGHV genes and devoid of both del11q and del17p who maximally benefit from such treatment; intermediate-risk patients harboring unmutated IGHV genes and/or del11q in the absence of del17p are a case mix of good and poor responders to FCR; while high-risk patients harboring del17p are unsuitable for chemoimmunotherapy. This model fails to consider the impact of recurrent CLL mutations of potential prognostic relevance, which may be improved by their inclusion. Purpose. We aimed at refining the genetic-based stratification of FCR-treated CLL patients by integrating the mutational profile in a prognostic model together with the IGHV mutation status and FISH karyotype. Methods. A multicenter cohort of 173 (162 with complete molecular data) untreated CLL receiving first-line therapy with FCR in the real-life clinical practice was evaluated by target resequencing. Tumor genomic DNA collected at the time of treatment was analyzed for mutations in the coding exons plus splice sites of CLL cancer driver genes (n=23). Deep next-generation-sequencing (NGS) of the gene panel was performed on the Illumina MiSeq platform (coverage >2000x in >90% of the target). Non-synonymous mutations represented in >10% of tumor allele were called by using VarScan2, and a stringent bioinformatic pipeline was developed to protect against the false call of polymorphisms and sequencing errors. Medical statistics was performed using SPSS version 24.0 and R version 3.3.2. Results. The cohort characteristics and mutational profile were consistent with those reported in CLL receiving FCR as initial treatment. After a median follow-up of 7.2 years, 114 patients progressed, accounting for a median PFS of 4.5 years. Among patients categorized as low-risk by IGHV and FISH status, gene mutations associated with poor prognosis were virtually absent (Fig. 1A). By univariate analysis, none of the cancer driver gene mutations significantly associated with PFS among low-risk patients. Consistently, by recursive partitioning, the proportion of low-risk patients failing FCR early was not explained by the co-occurrence of a cancer driver gene mutation. Among patients categorized as intermediate-risk by IGHV and FISH status, mutations of BIRC3 (HR: 6.452; 95% CI 2.467-16.875; p<0.001), BRAF (HR: 4.392; 95% CI 1.362-14.165; p=0.013), SAMHD1 (HR: 9.480; 95% CI 1.213-74.066; p=0.032), and ATM (HR: 2.282; 95% CI 1.028-5.068; p=0.043) associated with an increased risk of progression by univariate analysis. By multivariate analysis, mutations of BIRC3 (HR: 6.264; 95% CI 2.289-17.145; p<0.001) and BRAF (HR: 4.898; 95% CI: 1.493-16.071; p=0.009) maintained independent association with an increased risk of progression. Consistently, intermediate-risk patients according to IGHV and FISH were further stratified by recursive partitioning in two groups represented by those at high risk of failing FCR because of the presence of BIRC3 or BRAF mutations, and those wild type for both genes. This information helped refining our previous stratification model of FCR-treated patients based on CLL molecular features. Two high-risk groups emerged that shared a similarly poor PFS, namely i) TP53 mutated and/or deleted CLL (median PFS 2.1 years) and ii) BIRC3 or BRAF mutated CLL (median PFS 0.3 years), representing 4.9% of FCR-treated patients (Fig. 1B). Conversely, IGHV unmutated patients lacking alterations of TP53, BIRC3 and BRAF lesions had an intermediate outcome (median PFS 5.3 years). Patients with mutated IGHV genes, and lacking TP53, BIRC3, BRAF and del11q lesions, had an excellent outcome (median PFS not reached, 50.0% being progression-free at 10 years) (Fig. 1B). Conclusions. BIRC3 and BRAF mutations identify a very poor prognostic subgroup with wild type TP53 but failing FCR as cases harboring TP53 disruption. If validated, mutations of BIRC3 and BRAF might be used as molecular predictors to select high-risk patients for novel therapeutic approaches.
Forconi: AbbVie: Consultancy, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Infinity: Consultancy, Membership on an entity's Board of Directors or advisory committees; Gilead: Research Funding; Janssen-Cilag: Speakers Bureau. Zaja: Novartis: Honoraria, Research Funding; Janssem: Honoraria; Roche: Honoraria, Research Funding; Celgene: Honoraria, Research Funding; Abbvie: Honoraria; Takeda: Honoraria; Gilead: Honoraria. Rigolin: Celgene: Honoraria, Other: Advisory Board; Mundipharma: Other: Advisory Board; Gilead: Honoraria. Coscia: Abbvie: Membership on an entity's Board of Directors or advisory committees; Janssen: Honoraria, Research Funding; Gilead: Honoraria, Membership on an entity's Board of Directors or advisory committees. Tedeschi: AbbVie: Consultancy; Gilead: Consultancy, Other: travel expenses; Janssen: Consultancy, Other: travel expenses. Laurenti: Roche: Other: Advisory Board; Gilead: Other: Advisory Board; Janssen: Other: Advisory Board; Abbvie: Other: Advisory Board. Cuneo: Gilead: Honoraria, Other: Advisory Board; Janssen: Honoraria, Other: Advisory Board; Abbvie: Honoraria, Other: Advisory Board; Roche: Honoraria, Other: Advisory Board. Foa: AbbVie: Consultancy, Speakers Bureau; Novartis: Consultancy, Speakers Bureau; Sandoz: Consultancy, Speakers Bureau; BMS: Consultancy, Speakers Bureau; Celgene: Consultancy, Speakers Bureau; Amgen: Consultancy, Speakers Bureau; Gilead: Consultancy, Speakers Bureau; janssen: Consultancy, Speakers Bureau; Roche: Consultancy, Speakers Bureau. Gaidano: Gilead: Consultancy, Honoraria; Roche: Consultancy, Honoraria; Amgen: Consultancy, Honoraria; AbbVie: Consultancy, Honoraria; Janssen: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal